Authors: Leire Jauregi, Lur Epelde, Itziar Alkorta, Carlos Garbisu
Journal: Frontiers in Microbiology
Date: 2021
https://doi.org/10.3389/fmicb.2021.666854
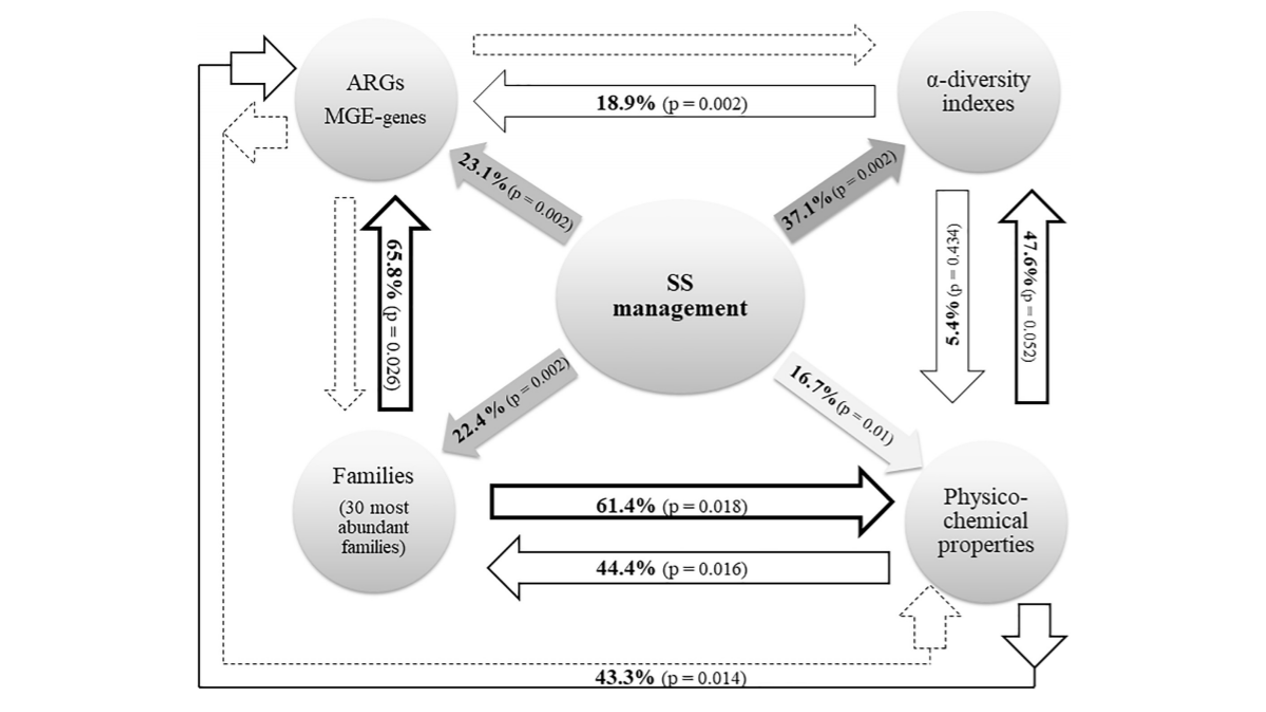
The application of sewage sludge (SS) to agricultural soil can help meet crop nutrient requirements and enhance soil properties, while reusing an organic by-product. However, SS can be a source of antibiotic resistance genes (ARGs) and mobile genetic elements (MGEs), resulting in an increased risk of antibiotic resistance dissemination. We studied the effect of the application of thermally-dried anaerobically-digested SS on (i) soil physicochemical and microbial properties, and (ii) the relative abundance of 85 ARGs and 10 MGE-genes in soil. Soil samples were taken from a variety of SS-amended agricultural fields differing in three factors: dose of application, dosage of application, and elapsed time after the last application. The relative abundance of both ARGs and MGE-genes was higher in SS-amended soils, compared to non-amended soils, particularly in those with a more recent SS application. Some physicochemical parameters (i.e., cation exchange capacity, copper concentration, phosphorus content) were positively correlated with the relative abundance of ARGs and MGE-genes. Sewage sludge application was the key factor to explain the distribution pattern of ARGs and MGE-genes. The 30 most abundant families within the soil prokaryotic community accounted for 66% of the total variation of ARG and MGE-gene relative abundances. Soil prokaryotic α-diversity was negatively correlated with the relative abundance of ARGs and MGE-genes. We concluded that agricultural soils amended with thermally-dried anaerobically-digested sewage sludge showed increased risk of antibiotic resistance dissemination.